Abstract
Background: Monitoring of BCR-ABL1 levels by quantitative PCR (qPCR) is essential for the management of CML patients treated with TKIs. Because of intrinsic limits, qPCR does not appear to be an optimal tool to quantify transcript levels around 10% and very low levels of disease. Digital PCR (dPCR), thanks to its intrinsic features, is expected to be more accurate than qPCR in the assessment of molecular MRD as demonstrated in different studies (Huang et al 2015, Franke et al, 2015). In order to hypothesize the use of this new technology in the clinical practice, a standardization process is mandatory.
Aim: We started a multicentric study in order to evaluate if different digital PCR instruments and methods can provide aligned results. For this purpose we analyzed the intra-assay reproducibility for each laboratory by calculating the % of coefficient variation (CV%).
Methods: Four laboratories were involved in the project: 3 have used a Bio-Rad instrumentation based on a droplet system (1 labs QX200, 2 lab QX100) and 1 used a chip-based instrumentation from Life Technologies (QuantStudio 3D). The assays used were: DigiDrop P210 MasterMix from Bioclarma s.r.l. (2 laboratories), "custom system" (2 laboratories). Tests were performed on cDNAs centrally prepared by one of the laboratory, over a period of 3 weeks. The same materials have been quantified by RT-QPCR to express the result in I.S. and dispatched to the other laboratories involved in the study. 16 RNA pools were prepared at different levels of disease (3 pools for 10% level, 3 pools for 1% level, 3 pools for 0.1% level, 3 pools for 0.01% level, 3 undetectable pools in MR 5.0, 1 pool of donors). Blind digital PCR analysis were performed and repeated in 3 separate sessions. ddPCR was performed in triplicate for BCR-ABL1 using 200 ng of cDNA for each replicate and in duplicate for ABL1 using 100 ng of cDNA for each replicate; the QuantStudio 3D was performed in duplicate for both genes using 50 ng of cDNA for each replicate of BCR-ABL1 and 25 ng of cDNA for each replicate of ABL1 (to avoid the saturation of the control gene).
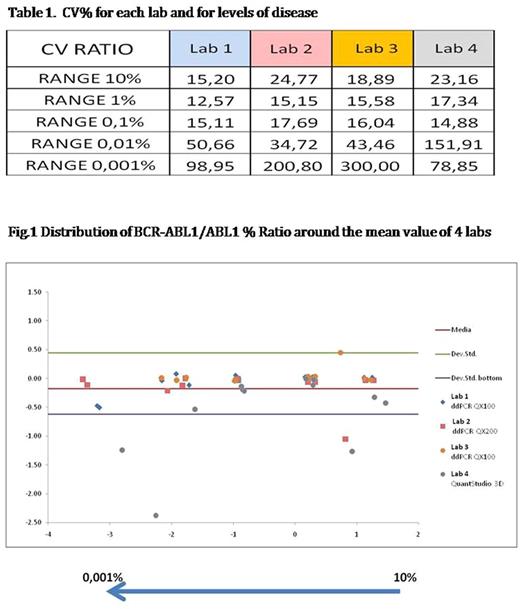
Results: The analysis was based on the data from the 4 laboratories by evaluating both of absolute copies of BCR-ABL1 and ratio (BCR-ABL1/ABL%). The analysis of intra-assay data for each sample in the three sessions and for each center showed that: the QuantStudio 3D system shows an overestimation of the values in all the levels of disease analyzed; 3 laboratories revealed 15% of positivity in the healthy samples (4/27 replicates); the laboratory with a chip-based approach didn't reveal any positivity but it tested only 50 ng of cDNA for each replicates (versus 200 ng in ddPCR) 2 out of 3 samples considered undetectable MR 5.0 in RT-qPCR resulted positive in 3 out of 4 laboratories. Then, for each sample we investigated how results obtained (ratio% and absolute copies of BCR-ABL1) for each sample were distributed around the median value calculated from all laboratories. Outlier values were excluded from the analysis according to Critical values for Grubbs outlier test. [Grubbs F: Technometrics 1969; 11(1):1-21]. The analysis showed that when expressed in ratio results of 3 out of 4 labs using ddPCR were within the range of ±2SD, the lab using the QuantStudio 3D had several values outside the ±2SD limit; regarding the absolute copies of BCR-ABL1 all values obtained, except for 2, were aligned around the mean value and fall within the range of the ±2SD. (Fig. 1) Finally, CV% calculated on the results (ratio% and absolute copies of BCR-ABL1) produced in 3 sessions of dPCR in each laboratory were similar for each lab for all levels of disease (Tab.1)
Conclusion: This multicentric Italian experience has highlighted a good overlapping of final data in terms of accuracy and reproducibility at different levels of disease, despite the presence of different sources of methodological and instrumental variability. Our results may suggest an improvement on variability respect to those obtained in RT-qPCR (Branford S. et al 2006). Despite the presence of different sources of methodological and instrumental variability, there is a good alignment of multicentric results.
Fava: Pfizer: Consultancy, Honoraria; Incyte: Consultancy, Honoraria; Novartis: Consultancy, Honoraria. Varotto: Bioclarma s.r.l.: Employment. Barberio: Bioclarma s.r.l.: Equity Ownership. Saglio: BMS: Consultancy, Honoraria; Ariad: Consultancy, Honoraria; Incyte: Consultancy, Honoraria; Roche: Consultancy, Honoraria; Incyte: Consultancy, Honoraria; Novartis: Consultancy, Honoraria; Ariad: Consultancy, Honoraria; Pfizer: Consultancy, Honoraria; Roche: Consultancy, Honoraria; BMS: Consultancy, Honoraria; Pfizer: Consultancy, Honoraria; Novartis: Consultancy, Honoraria. Pane: Novartis: Honoraria, Speakers Bureau. Martinelli: PFIZER: Consultancy; ARIAD/INCYTE: Consultancy; AMGEN: Consultancy; ROCHE: Consultancy; JOHNSON & JOHNSON: Consultancy; CELGENE: Consultancy. Specchia: Novartis: Consultancy, Honoraria; BMS: Consultancy, Honoraria; Incyte: Consultancy, Honoraria; Pfizer: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal